Since the turn of the century, every decade has witnessed the development of new high-throughput technologies to monitor diverse layers of biological processes (e.g., genome, transcriptome, proteome, interactome, metabolome). Each of these technologies motivated the development of new bioinformatics approaches and tools to extract relevant information from the raw data. However, an integrated approach is still lagging behind to translate this diversity of complex and heterogeneous data into useful knowledge.
The IFB’s innovation axis aims to build a strategic vision to ensure the consistency and relevance of national bioinformatics developments and services, as well as their links with European initiatives. A major challenge is to develop tools and services to address a very important challenge of today’s biology: the integration of heterogeneous high-throughput data, in order to approach the complexity of biological systems on a holistic scale. A number of networks and national projects dedicated to the production, management and interpretation of specific data types (genomics, proteomics, metabolomics, imaging, animal models, cohorts, personalised medicine etc.) have been set up in France. In order to assume a global view of the systems which are studied, this heterogeneous information need to be integrated.
We, therefore, proposed to tackle the challenge of integrative bioinformatics through three actions of the new action plan:
In September 2018, Aviesan ITMO Genetics, Genomics and Bioinformatics (GGB) and IFB coorganised a one-day workshop titled Challenges and Perspectives in Integrative Bioinformatics, which gathered 80 attendees. A series of talks presented the state-of-the-art approaches in integrative bioinformatics (multilevel matrix factorisation methods and multilayer network analysis). Overall, the meeting covered all the steps between data production, computational treatment, statistical analysis, visualisation and interpretation, illustrated by relevant examples from various domains of life sciences and their applications. The talk sessions were followed by a round-table session where all participants discussed the personal experience, current needs and problems encountered, which led to an animated discussion about current challenges, perspectives and strategies to further develop integrative bioinformatics. Beyond the scientific interest of the talks and debate, this one-day workshop also enabled us to identify experts in the field, who were then invited, and accepted, to contribute to the University Diploma in Integrative Bioinformatics
The IFB Action Plan 2018-2021 includes an innovative component focusing on integratrive bioinformatics (WP2), aimed at ensuring the coherence and relevance of bioinformatics developments and services and their link with European initiatives (ELIXIR, inter-ESFRI call). In this context, the major challenge, with these pilot projects, is to stimulate the development of tools and services to integrate heterogeneous data at high throughput (sequencing, imaging, proteomics, metabolomics, etc.) in order to first address the complexity of biological systems. The relevance of these approaches must be demonstrated by applying them to specific use cases based on data generated by at least two national research infrastructures, animal model facilities, human cohorts or national plans.
Projects must also be in line with national priorities, including health, food safety and security, the environment, biotechnology and basic research. The pilot projects have been designed to achieve results in the relatively short term by building on the existing know-how of different IAP structures (infrastructures, cohorts, plans). The call for LOI was formulated as follows: within a period of 18-24 months, to answer a specific research question using an integrative bioinformatics approach illustrated by specific case study involving several national infrastructures. Successful applications were supported by a fixed-term contract of up to two years.
The pilot projects involve ten national research infrastructures (IFB, France Génomique, MetaboHub, ProFI, FRISBI, FBI, FLI, Phenome, EMBRC-Fr, F-CRIN), two model facilities for animals and human cohorts (PHENOMIN, BIOBANQUES) and the national plan "France Médecine Génomique 2025".
Five projects were selected:
In modern medicine, the combination of genomic, clinical and imaging data is rapidly becoming more widespread for diagnosis and therapeutic decision-making. The aim of the INEX-MED project is to develop a perennial infrastructure for the integration, exploitation and modelling (semantic/statistical) on heterogeneous data sources and multi-site computing infrastructures in the biomedical field. The technological developments, around (i) the construction of knowledge bases adopting the principles of "FAIR data" (Findability, Accessibility, Interoperability, Reusability), (ii) the interrogation of secured data, and (iii) the integrated exploration of data via bio-statistical and automatic learning methods, will be shared between the partners. This infrastructure will then be exploited and validated through integrative approaches in order to improve diagnosis/prognosis in two application scenarios: (i) INGEN-IA, for the study of the formation and development of intracranial aneurysms, and (ii) MYO-lico, for assistance in the diagnosis of different forms of myopathies. Emphasis will be placed on the creation of a generic, modular and portable infrastructure that can be installed on IFB resources and reused by the entire biomedical community.
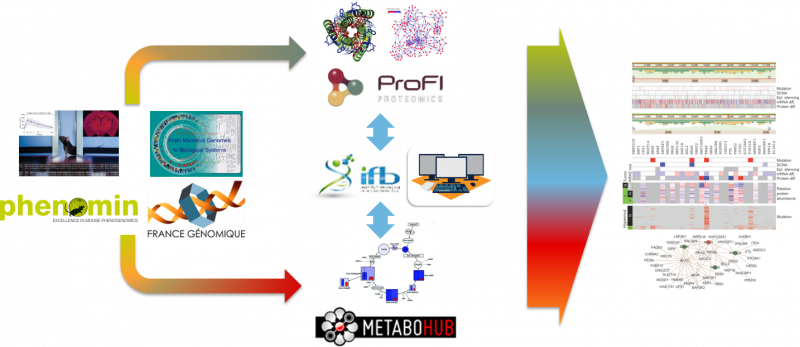
Proteomics and metabolomics approaches provide unique and complementary information to decipher gene function, elucidate phenotypes, and robustly discover new biomarkers for disease treatments. National bioinformatics resources have been recently developed by the IFB, ProFI, and MetaboHUB infrastructures to analyse each of these omics data. Here, we propose to develop new tools enabling high-throughput and combined proteomics and metabolomics data analysis. First, statistical modeling will be used to explore the specific and common information from the various data, and to determine how their combination as molecular signatures can optimally interpret and predict the phenotypes. Second, network integration will be used to facilitate interpretation and annotation of the data. The mathematical approaches and tools will be validated on two use cases focusing respectively on high-throughput phenotyping of mouse models generated by the PHENOMIN infrastructure, and the discovery of new gene functions in systems microbiology approaches developed by France Genomique. The project will provide the national Workflow4Metabolomics bioinformatics resource with innovative tools and workflows for high-throughput combined proteomics and metabolomics data analysis, and pave the way for new phenotyping services provided to academia and industry. It will also enrich the PHENOMIN IMPC database of mouse models and the MicroScope microbial knowledge base.
Our project is primarily based on the Nucleipark longitudinal study, which aims to identify new biomarkers for the diagnosis and progression of idiopathic Parkinson's disease, as well as two complementary studies (HTP and PREDICTOR), relating to portal hypertension and cancers of the upper aerodigestive tract. Nucleipark is based on the acquisition of a wide variety of data: clinical information, multimodal neuroimaging, transcriptomics, methylomics, metabolomics and lipidomics. HTP and PREDICTOR involve radiomic analyses (extraction of a large number of quantitative parameters from in vivo images) and their links with biological and omic data, in order to obtain better diagnostic and prognostic indicators. When dealing with cohorts of subjects characterized by heterogeneous information, of large dimensions, and measured over time, their interpretation, and in particular the identification of multimodal biomarkers, requires the use of adapted statistical integration methods, such as Regularized Generalized Generalized Canonical Analysis (RGCCA) and its extensions. Currently, an R package is available for "expert" command line use of these methodologies. We would therefore like to propose access modalities adapted to a biologist or clinician end-user, through the development of ergonomic and intuitive graphical interfaces.
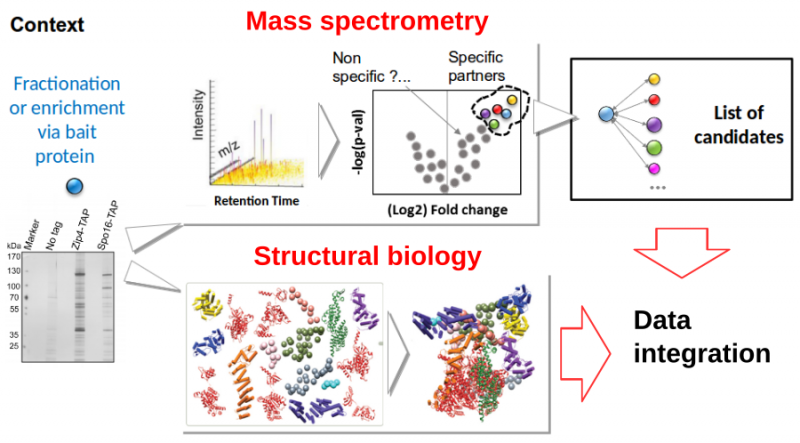
Mass spectrometry is now an essential technique for the characterization of molecular species and their interactions. It has recently acquired extreme accuracy and high throughput while requiring little equipment. For many biological questions, proteomics or interactomics analyses are now essential entry points. However, these studies often stop at the step of enumerating interacting macromolecules (mainly proteins) without further analysis of the identified sequences. However, structural or evolutionary aspects offer a powerful interpretation grid for biologists (interpretation of mutations in patients disrupting assemblies, targeted mutagenesis and functional dissection, virtual screening, etc.). We propose to bring together and combine high-throughput bioinformatics tools for the modeling of protein structures and their assemblages. The service deployed will enable PROFI users to facilitate post-processing of the screens and amplify the exploitable information. At the same time, it will benefit FRISBI users for the integrative modelling of data acquired on their platforms. The coupling between these two major strategies for interactome characterization will be enhanced by the bioinformatics environment offered by the IFB. It will benefit the entire community of biologists working on interactions between macromolecules improving, for example, the analysis of pathological dysfunctions (modification of interactions, isoforms, etc.).
Phenome or Emphasis-fr, the national infrastructure for plant phenotyping, has nine nodes, or experimental platforms, spread over the national territory. Each of these nodes has an information system, in particular PHIS, which is currently in production on two nodes and being deployed on others. One of these nodes, HiTMe, supports the metabolic phenotyping of samples taken from the other nodes. It is also one of the 4 components of the Metabolome Bordeaux platform, itself part of the MetaboHub infrastructure. The integration of the data produced by these different nodes is based on the standards built jointly by Emphasis and Elixir. It involves in particular the definition of ontologies of phenotypic and environmental variables, the definition of the MIAPPE standard and its implementation via the Breeding API web services implemented in PHIS and in GnpIS, the INRA information system belonging to Elixir-fr that allows the publication of plant phenotyping data. The objective of this project will be to enable the consolidation and integration of different corn data sets, obtained through several scientific projects, in order to explore the reciprocal impact of environmental conditions (cultivation practices, pedoclimatic conditions, ...) and the choice of corn varieties on the synthesis of different metabolites of interest.
Published Datasets
Publication related to the project
Other contribution
Three additional projects have been reoriented to be supported by other IFB actions: "Support to databases", "Catalysing interoperability" and "Sharing services with national research infrastructures":
Holders : Faisal Bekkouche (EMBRC, Villefranche/mer)
AIP structures : EMBRC, FBI
EMBRC-En/EU is an infrastructure of excellence providing access to marine living resources, their ecosystems, and techniques for analysing these marine resources for any public/private user worldwide. The French node of EMBRC-ERIC offers high-tech marine organism imaging strategies (live imaging, 3D imaging and high-speed imaging) on the three sites of the infrastructure (Institut de la Mer de Villefranche, Observatoire Océanologique de Banyuls and Station Biologique de Roscoff). Recent progress in the field of imaging has led to an exponential increase in the amount of information collected by the data-producing centres. The optimal exploitation of this data is essential to unlock the application of marine models to numerous fields of research and application, as well as for the exploitation and preservation of ecosystems and marine biodiversity. The implementation of the My-EMBRC-IMAGE webservice, combined with a distributed storage infrastructure accessible remotely, enables the saving, management, visualisation and analysis of microscopy images. This project, based on a synergy between the EMBRC-France, FBI and IFB infrastructures, should increase their influence and impact on the scientific communities and the public.
Holders : Pascal Roy (IFB PRABI, Lyon)
AIP structures : France Génomique, F-CRIN
The project is part of the IFB's 2018-2021 programme, more specifically responding to the objective of integrating resources and strengthening interoperability between teams. This structuring project consists of setting up a network of expert teams for the statistical analysis of large-scale data and the contextualised interpretation of the results of the analysis of the massive parallel sequencing of the human genome and infectious agents. The project will contribute to the implementation of the statistical analysis methods developed by the partner teams on the different platforms on the one hand, and to the development and validation of new statistical analysis methods within the framework of collaborations between these teams on the other hand. The project concerns the statistical analysis of the performances of sequencing pipelines, the study of the properties of identification models of diagnostic, prognostic and theranostic biomarkers, and the development of models integrating the biological and experimental components of variability. Emphasis is placed on statistical inference and the study of the predictive properties of models in the context of their applications in epidemiology and clinical medicine. The structuring action combines the dissemination of methods and the setting up of collaborative projects between platforms.
Holders : Christophe Beroud & David Salgado (Inserm, Marseille)
AIP structures : -
Within the framework of the national network for the diagnosis of genetic diseases, the Plan France Médecine Génomique 2025 and the ELIXIR Human Data community, the collection and standardised annotation of constitutional Copy Number Variations (CNVs) found in patients is of major interest to facilitate the identification of new genes responsible for genetic diseases (research), the identification of pathogenic CNVs and to facilitate medical interpretation with a reduction in the number of Variant of Unknown Significance (VUS) (diagnosis). France, via the GMGF-GBIT platform in Marseille, is the leader of the "Human CNV" community of ELIXIR and is developing the national BANCCO database in partnership with the national network of AchroPuce diagnostic laboratories, which brings together all the laboratories carrying out Chromosomal Analysis on DNA chips (ACPA or CGH array) in a diagnostic framework for constitutional pathologies. This development is being carried out with the Hospices Civils de Lyon team led by Professor Sanlaville, who is also co-responsible for the AURAGEN platform of the France Genomic Medicine 2025 Plan. Professor Christophe Béroud (GMGF-GBIT) is also involved in the Plan France Médecine Génomique 2025 where he is one of the experts of the CAD (Data Collector and Analyser) committee. To date, the beta version of BANCCO contains data from 17,000 patients and more than 35,000 CNVs and will be rapidly enriched. The collection of heterogeneous data from external databases, preaching tools and expert annotations will also make it possible to offer the scientific community diagnostic quality data enriched with numerous annotations and classification.